Where Can I Upload Plink Binaries for Analysis
Your showtime PLINK tutorial
Learning outcomes: At the end of this chapter, you will be able to modify genotype information formats with PLINK.
In the previous posts, you read almost the full general suggestions for the work environment, downloaded the PLINK software, and genotype data for a surprisingly big number of animals. But the program was non executed yet in any meaningful way... But now everything will change and y'all will finally run the PLINK programme. Your goal will be to transform the binary file format saved as bim, bed, and fam files to a text-based genotype format saved every bit ped and map files. This exercise will also give y'all a detailed clarification of the use of PLINK. You volition see that it is not difficult at all. Frankly, I look at the program as some kind of building-cake game. You need to know what you want to achieve and all you need to practice is add the correct elements to it. The start matter y'all need to write downwardly is some sort of base structure. In fact, you can first with this very same line all the fourth dimension and add elements to information technology.
But earlier you begin...
...I know, I know I am abrasive with these recommendations all the fourth dimension, and you are eager to jump in... But hear me out... Y'all can type the PLINK commands directly to the command line, but don't do that. You lot volition see that there volition exist many mistakes and re-runs all the time, and this way you will need to re-type all the time. This is a huge loss of time. You don't want that. Open a new text file instead and write your plan script in that location. Ideally, this text file is saved in a cloud storage directory, then it is being automatically backupped upon relieve. Call back: the script files are very pocket-size in size, merely extremely valuable given the corporeality of time yous invested in writing them.
...1 more piece of advice, in case you are new to scripting and programming. You lot might be surprised, but the scripts you write should be readable by the computer, but perhaps fifty-fifty more importantly by people, including future y'all. Allow me explain... You write whatever script today and you sort of know what it does. I guarantee y'all that if you lot come up back to it even after a week, you will have to spend quite some time figuring out what it does. Not to mention if you wrote some stuff like two years ago... Or imagine that y'all have to send this script to your colleague, who was not involved in the writing at all! If it is not clear how to change fifty-fifty bones things like input file names or locations, you lot are merely looking for bug. And then only document your code using plain words what some crucial lines or sections do. I use the # hashtag sign at the beginning of each comment line to indicate it equally such. Also, lines starting with a # are ignored in many programs, so exercise non cause general errors.
Long story short: Document your code!
So now yous will run PLINK. For real this time... Open the command prompt in a folder where you have the plink executable file and the genotype information, as described before in the PLINK - Software for genomic analyses chapter. Open up a new text file and re-create the post-obit lines in there:
# Change binary genotype to ped+map format plink --bfile ADAPTmap_genotypeTOP_20160222_full --cow --nonfounders --let-no-sexual activity --recode --out ADAPTmap_TOP Relieve the text file. From now on any change you implement volition be written to the text file commencement, then you can adapt easily in instance of need. Copy the whole plink line to the command prompt (without the annotate line) and press enter. You take to accept 1Gb complimentary space for the recoded file. If everything went well, you lot will encounter this:
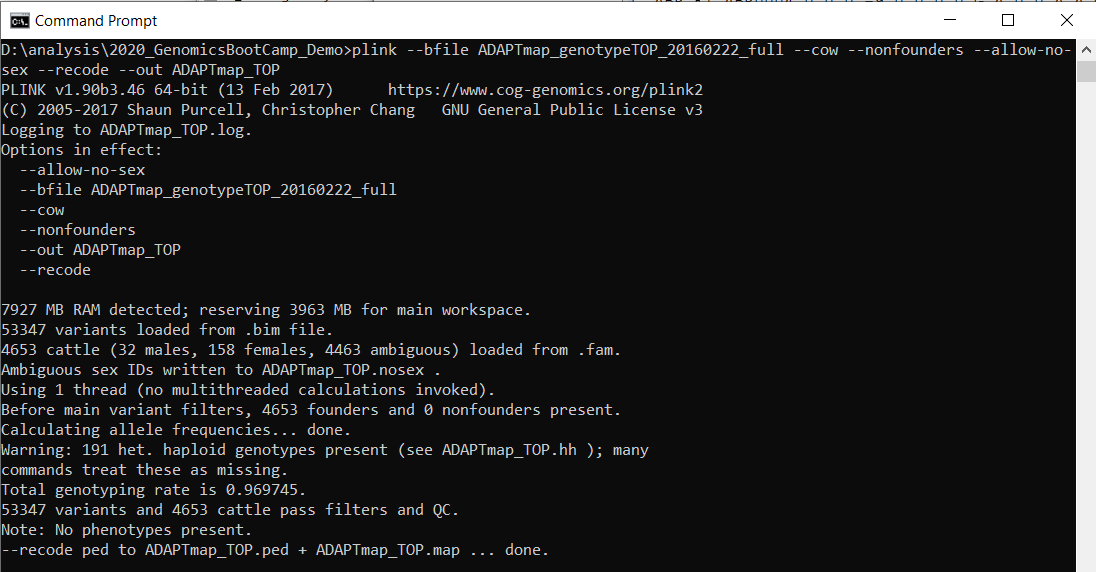
Effigy seven.1: A successful PLINK run
In the following section I volition explicate what yous just did in ii parts:
Start, allow's start with the PLINK options. I will list them, simultaneously providing a link to them on the PLINK website. I will also tell yous how can you (hands?) discover answers for any PLINK option.
2nd, I volition talk about the resulting ped and map files, including their structure.
The PLINK options
I beginning with a general annotate about the overall structure of PLINK that you take already noticed. Subsequently the program name, there are various options preceded past a double dash "--". Some less-appropriate-text-editors like MS Word might autocorrect this to a long dash, which will effect in an mistake. So again, simply use a proper text editor.
The PLINK options come in two formats:
--optinName1
--optionName2 space additionalParameter(south)RelatedToOptinName2
What the options used in the previous run mean:
--bfile ADAPTmap_genotypeTOP_20160222_full This is how yous specify that your input file format is a binary ped file. Because you take all 3 files in the same directory as the PLINK executable, you simply need to specify the file prefix, and the program will automatically utilise everything from the bim, bed, and fam files.
--cow This specifies the number of chromosomes in your data set. In instance y'all practice non tell anything about chromosome sets, the programme volition utilise the homo genome as a default setting. Now yous might have noticed that nosotros bargain with goats, simply we specify --cow hither. This time nosotros played on the fact that bot cattle and goats have 60 chromosomes. In instance yous employ another organism, you lot either specify that option, or a new setting using --chr-ready
--nonfounders Exercise you remember when I talked almost parental data in the fam file? This choice is related to bypass whatsoever animals with missing parent information existence treated as founders. At present, when y'all read the description on the website, you volition likewise come across that it is considered simply in a few cases anyway. The affair is, however, that I tend to forget to utilize the handle in specific cases, and so spend a lot of fourth dimension figuring out what went incorrect. This is the pesky type of error when there is no error message, but the outcome is just wrong. So to avoid all of this, I use --nonfounders in all my plink lines.
--let-no-sex Similar to parental info, the info on sexual activity is also many times missing. Equally it might or might not be problematic for certain analyses, I have decided to include this option in all my PLINK lines. The justification is the same as for --nonfounders.
--recode This very simple command is the i that actually does all the work. Past putting this in your lawmaking y'all specify that you want to have ped and map files every bit output.
--out ADAPTmap_TOP Specifies the file name for newly created files.
So at present you might accept a very valid question: "This is all nice, only how do I find descriptions for other types of analyses I want to do?"
The ii possibilities are:
- If you know exactly what are you looking for, you tin can utilise the handy search tool at the lesser of the left console on the website, as shown in the picture. The result(s) beneath the search box are clickable links to the resource.
Figure 7.2: Find this bottom left on the PLINK website to go help
- If you don't know what you are looking for, try to explore the topics discussed on the left panel on the PLINK website, check other people's code, or only ask colleagues how to practice something. Alternatively, you can also attempt to fish for answers in the listing of all PLINK options, using Ctrl+F text search capacities of your browser. You lot can also get there by clicking the "index" keyword link above the search bar.
Here I would note that PLINK can practise many things, but certainly non everything. So practice non await to be a one-stop-shop for all of your genomic assay needs. It is just a tool. A handy one, but all the same just one out of many.
The ped and map file format
In this department, we will accept a brief wait at the newly created files and tell something nearly their structure.
You might have noticed that there are a few new files created in the same directory you accept run the program. From these files, the ones with file extension .ped and .map are the most important.
The .map file is very similar to the previously described .bim file, only without the concluding two columns with genotypes.
The .ped file structure is essentially the chain of the .fam file (one line per individual), followed by human-readable genotypes in text format. Every ii columns stand for one SNP in a infinite-delimited format.
To open up the .map file should be no problem. The .ped file however is nearly 1Gb in size! I got the "File as well large to be opened" error message with Notepad++, but the TextPad opened it without problems (subsequently a bit of waiting fourth dimension, might be auto-dependent, you lot will see a small progress bar lesser left).
How to run PLINK from R
Every bit a practical demonstration of work with genomic data in R Studio, nosotros will use PLINK example we discussed before in this chapter. With this, you will meet the elements that need to be included to integrate the PLINK script to R and too set you for the one thousand finale of the first section - the PCA assay.
The script nosotros will be running is the following:
# clear workspace rm(list = ls()) # set working directory setwd("d:/analysis/2020_GenomicsBootCamp_Demo/") # run PLINK QC system("plink --bfile ADAPTmap_genotypeTOP_20160222_full --cow --nonfounders --permit-no-sex --recode --out ADAPTmap_TOP") From my personal experience in learning and teaching genomic data analysis to people of wide-ranging levels of experience, simply telling you lot to re-create-paste-and-run-the-script does not atomic number 82 anywhere. Then I volition explicate the most important elements of this script.
All lines starting with a hashtag # are comment lines. As I explained in a previous mail service, these are essential in all scripts. You need to be very clear what each part of the script is doing, which is much easier if you comment on it. The line rm(list = ls()) is a very useful ane that I employ in all my R scripts. It deletes everything from the workspace. This style I know that in that location are no pesky leftovers from previous analyses that might compromise my runs. (Ever had an experience of "The script was running earlier and it is not running now!"? This might be considering of unintended information sets in the work environment.)
The setwd() is an R function that (equally the name implies) sets the working directory for this session. The working directory is the place, where R volition look for all the data for the analyses and place any output files if you do not specify otherwise. Past default, this is a directory somewhere on your system drive. While information technology volition piece of work, I strongly advise changing it to comply with your ain file organization structure in a custom directory, equally discussed before. You will need to modify this for your run. Simply put the PATH to your working directory containing PLINK and the genotype files between the quotation marks. Please note that R uses the opposite slashes as Windows.
You probably recognize the contents of the system() part. This is the exact copy of the PLINK command we used earlier, again between quotation marks. That's right! It is this easy to run PLINK from R. With the use of this role, the R opens the system command line, runs the line of code, and closes the control line.
Each line of the script above could be run separately. To do this, just put your cursor to the line y'all want to execute, and press the "Run" push in the top right corner. Alternatively, you can use the even better and arguably quicker method of simultaneously pressing Ctrl+Enter (Command+Enter in Mac).
Exercise
Phewww... Y'all made information technology to the end of this unexpectedly long clarification! Congratulations! But to really bulldoze domicile the message and lock the noesis in your memory, I have a small job for you.
You see, the PLINK file formats are actually popular, but at that place are many others out in that location. The good news is, that you tin use PLINK to transform files to other popular formats. 1 of them is undoubtedly the so-chosen variant telephone call format that is the standard output file from whole-genome sequencing pipelines, and a possible input to some other programs. So your task is to change the ADAPTmap file to vcf file format.
Hint: if I were you lot, I would explore the diverse options of the --recode choice on the website. wink-flash
As always, you can compare your solution to the one on YouTube. The video also contains some bonus information on related problems you might face during analyses, then brand sure to check it out regardless.
If the embedded video does not get-go, click it over again to "Watch on YouTube". Direct link: https://world wide web.youtube.com/spotter?v=c1LSFiv9CxY
Source: https://genomicsbootcamp.github.io/book/your-first-plink-tutorial.html
0 Response to "Where Can I Upload Plink Binaries for Analysis"
Post a Comment